mirror of
https://github.com/nod-ai/AMD-SHARK-Studio.git
synced 2026-02-19 11:56:43 -05:00
119 lines
6.9 KiB
Markdown
119 lines
6.9 KiB
Markdown
# Overview
|
|
|
|
This document is intended to provide a starting point for profiling with SHARK/IREE. At it's core
|
|
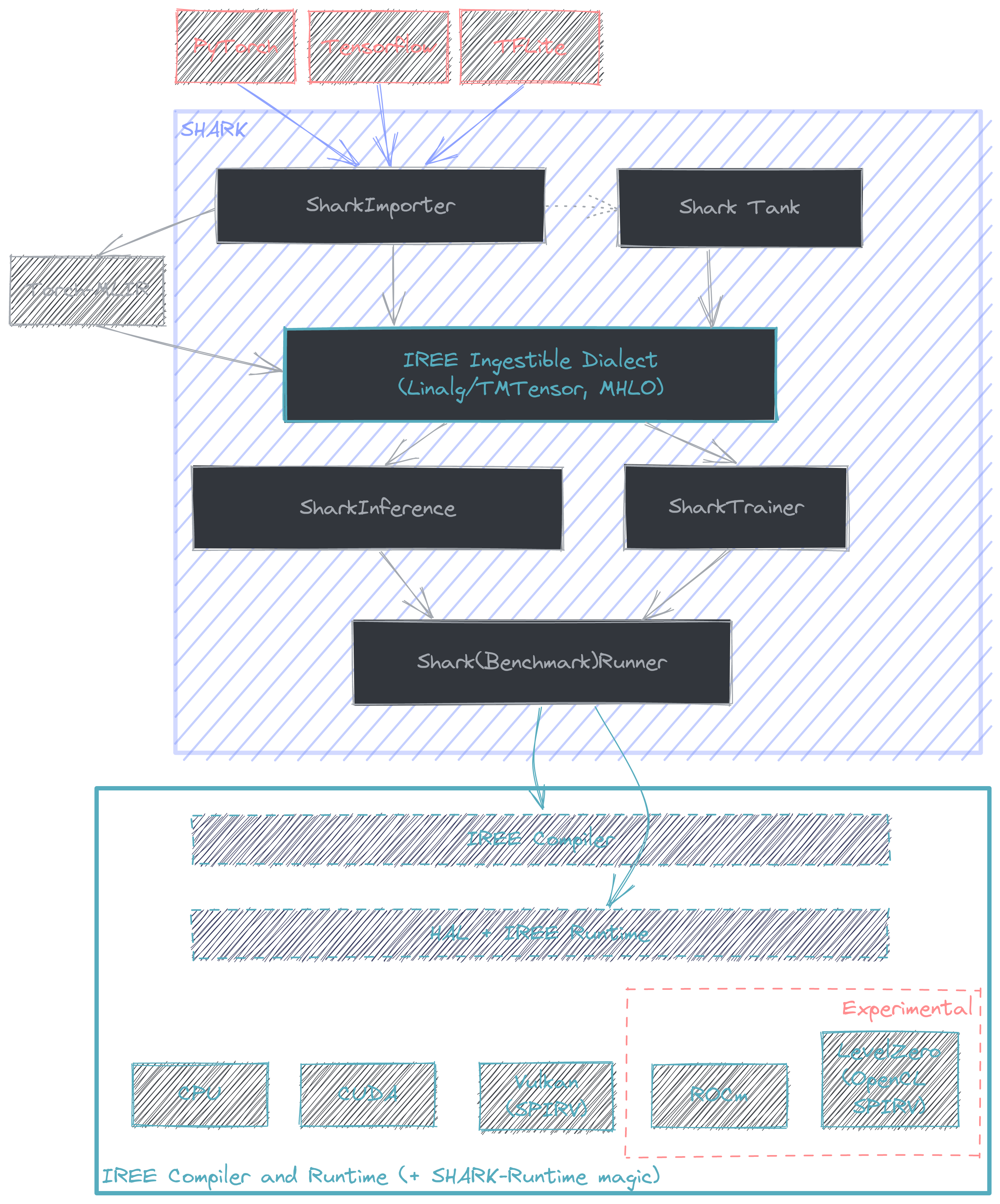
[SHARK](https://github.com/nod-ai/SHARK/tree/main/tank) is a python API that links the MLIR lowerings from various
|
|
frameworks + frontends (e.g. PyTorch -> Torch-MLIR) with the compiler + runtime offered by IREE. More information
|
|
on model coverage and framework support can be found [here](https://github.com/nod-ai/SHARK/tree/main/tank). The intended
|
|
use case for SHARK is for compilation and deployment of performant state of the art AI models.
|
|
|
|
|
|
|
|
## Benchmarking with SHARK
|
|
|
|
TODO: Expand this section.
|
|
|
|
SHARK offers native benchmarking support, although because it is model focused, fine grain profiling is
|
|
hidden when compared against the common "model benchmarking suite" use case SHARK is good at.
|
|
|
|
### SharkBenchmarkRunner
|
|
|
|
SharkBenchmarkRunner is a class designed for benchmarking models against other runtimes.
|
|
TODO: List supported runtimes for comparison + example on how to benchmark with it.
|
|
|
|
## Directly profiling IREE
|
|
|
|
A number of excellent developer resources on profiling with IREE can be
|
|
found [here](https://github.com/iree-org/iree/tree/main/docs/developers/developing_iree). As a result this section will
|
|
focus on the bridging the gap between the two.
|
|
- https://github.com/iree-org/iree/blob/main/docs/developers/developing_iree/profiling.md
|
|
- https://github.com/iree-org/iree/blob/main/docs/developers/developing_iree/profiling_with_tracy.md
|
|
- https://github.com/iree-org/iree/blob/main/docs/developers/developing_iree/profiling_vulkan_gpu.md
|
|
- https://github.com/iree-org/iree/blob/main/docs/developers/developing_iree/profiling_cpu_events.md
|
|
|
|
Internally, SHARK builds a pair of IREE commands to compile + run a model. At a high level the flow starts with the
|
|
model represented with a high level dialect (commonly Linalg) and is compiled to a flatbuffer (.vmfb) that
|
|
the runtime is capable of ingesting. At this point (with potentially a few runtime flags) the compiled model is then run
|
|
through the IREE runtime. This is all facilitated with the IREE python bindings, which offers a convenient method
|
|
to capture the compile command SHARK comes up with. This is done by setting the environment variable
|
|
`IREE_SAVE_TEMPS` to point to a directory of choice, e.g. for stable diffusion
|
|
```
|
|
# Linux
|
|
$ export IREE_SAVE_TEMPS=/path/to/some/directory
|
|
# Windows
|
|
$ $env:IREE_SAVE_TEMPS="C:\path\to\some\directory"
|
|
$ python apps/stable_diffusion/scripts/txt2img.py -p "a photograph of an astronaut riding a horse" --save_vmfb
|
|
```
|
|
NOTE: Currently this will only save the compile command + input MLIR for a single model if run in a pipeline.
|
|
In the case of stable diffusion this (should) be UNet so to get examples for other models in the pipeline they
|
|
need to be extracted and tested individually.
|
|
|
|
The save temps directory should contain three files: `core-command-line.txt`, `core-input.mlir`, and `core-output.bin`.
|
|
The command line for compilation will start something like this, where the `-` needs to be replaced with the path to `core-input.mlir`.
|
|
```
|
|
/home/quinn/nod/iree-build/compiler/bindings/python/iree/compiler/tools/../_mlir_libs/iree-compile - --iree-input-type=none ...
|
|
```
|
|
The `-o output_filename.vmfb` flag can be used to specify the location to save the compiled vmfb. Note that a dump of the
|
|
dispatches that can be compiled + run in isolation can be generated by adding `--iree-hal-dump-executable-benchmarks-to=/some/directory`. Say, if they are in the `benchmarks` directory, the following compile/run commands would work for Vulkan on RDNA3.
|
|
```
|
|
iree-compile --iree-input-type=none --iree-hal-target-backends=vulkan --iree-vulkan-target-triple=rdna3-unknown-linux benchmarks/module_forward_dispatch_${NUM}_vulkan_spirv_fb.mlir -o benchmarks/module_forward_dispatch_${NUM}_vulkan_spirv_fb.vmfb
|
|
|
|
iree-benchmark-module --module=benchmarks/module_forward_dispatch_${NUM}_vulkan_spirv_fb.vmfb --function=forward --device=vulkan
|
|
```
|
|
Where `${NUM}` is the dispatch number that you want to benchmark/profile in isolation.
|
|
|
|
### Enabling Tracy for Vulkan profiling
|
|
|
|
To begin profiling with Tracy, a build of IREE runtime with tracing enabled is needed. SHARK-Runtime (SRT) builds an
|
|
instrumented version alongside the normal version nightly (.whls typically found [here](https://github.com/nod-ai/SRT/releases)), however this is only available for Linux. For Windows, tracing can be enabled by enabling a CMake flag.
|
|
```
|
|
$env:IREE_ENABLE_RUNTIME_TRACING="ON"
|
|
```
|
|
Getting a trace can then be done by setting environment variable `TRACY_NO_EXIT=1` and running the program that is to be
|
|
traced. Then, to actually capture the trace, use the `iree-tracy-capture` tool in a different terminal. Note that to get
|
|
the capture and profiler tools the `IREE_BUILD_TRACY=ON` CMake flag needs to be set.
|
|
```
|
|
TRACY_NO_EXIT=1 python apps/stable_diffusion/scripts/txt2img.py -p "a photograph of an astronaut riding a horse"
|
|
|
|
# (in another terminal, either on the same machine or through ssh with a tunnel through port 8086)
|
|
iree-tracy-capture -o trace_filename.tracy
|
|
```
|
|
To do it over ssh, the flow looks like this
|
|
```
|
|
# From terminal 1 on local machine
|
|
ssh -L 8086:localhost:8086 <remote_server_name>
|
|
TRACY_NO_EXIT=1 python apps/stable_diffusion/scripts/txt2img.py -p "a photograph of an astronaut riding a horse"
|
|
|
|
# From terminal 2 on local machine. Requires having built IREE with the CMake flag `IREE_BUILD_TRACY=ON` to build the required tooling.
|
|
iree-tracy-capture -o /path/to/trace.tracy
|
|
```
|
|
|
|
The trace can then be viewed with
|
|
```
|
|
iree-tracy-profiler /path/to/trace.tracy
|
|
```
|
|
Capturing a runtime trace will work with any IREE tooling that uses the runtime. For example, `iree-benchmark-module`
|
|
can be used for benchmarking an individual module. Importantly this means that any SHARK script can be profiled with tracy.
|
|
|
|
NOTE: Not all backends have the same tracy support. This writeup is focused on CPU/Vulkan backends but there is recently added support for tracing on CUDA (requires the `--cuda_tracing` flag).
|
|
|
|
## Experimental RGP support
|
|
|
|
TODO: This section is temporary until proper RGP support is added.
|
|
|
|
Currently, for stable diffusion there is a flag for enabling UNet to be visible to RGP with `--enable_rgp`. To get a proper capture though, the `DevModeSqttPrepareFrameCount=1` flag needs to be set for the driver (done with `VkPanel` on Windows).
|
|
With these two settings, a single iteration of UNet can be captured.
|
|
|
|
(AMD only) To get a dump of the pipelines (result of compiled SPIR-V) the `EnablePipelineDump=1` driver flag can be set. The
|
|
files will typically be dumped to a directory called `spvPipeline` (on Linux `/var/tmp/spvPipeline`. The dumped files will
|
|
include header information that can be used to map back to the source dispatch/SPIR-V, e.g.
|
|
```
|
|
[Version]
|
|
version = 57
|
|
|
|
[CsSpvFile]
|
|
fileName = Shader_0x946C08DFD0C10D9A.spv
|
|
|
|
[CsInfo]
|
|
entryPoint = forward_dispatch_193_matmul_256x65536x2304
|
|
```
|